Epidemiología y genética del ganado lechero de pequeños productores.
Este proyecto postdoctoral tenía como objetivo investigar las enfermedades infecciosas que afectan al ganado lechero en pequeñas granjas de Tanzania, así como realizar estudios de asociaciones de todo el genoma para comprender la base genética de la respuesta de los anticuerpos.
6/16/20242 min leer


Smallholder dairy cattle in Tanzania
Smallholder farming is an important source of income for Tanzanian households. This project programme was part of the activities of the CTLGH phase 1 partnership between several international organisations, including the University of Edinburgh and the Gates Foundation.
Data Collection and Preparation
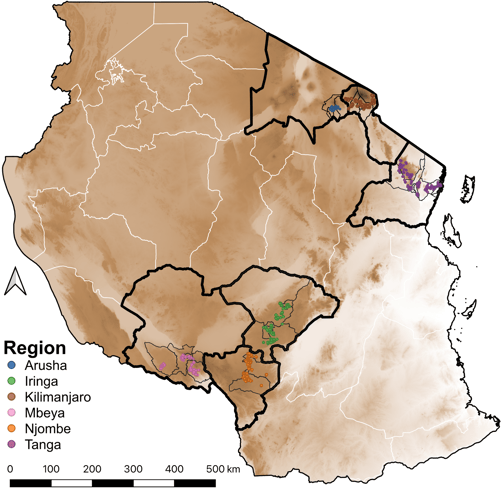
In this project, a team of MSc and PhD Tanzanian veterinarian students and assistants collected samples and surveyed hundreds of farms and households across six important regions for dairy production in Tanzania. The team also tested samples for antibody response to seven pathogens (Bovine viral diarrhoea virus (BVDV), Neospora caninum, Leptospira interrogans serovar Hardjo, Rift Valley Fever virus (RVFV), Toxoplasma gondii, Coxiella Burnetii, and Brucella abortus) commonly affecting dairy cattle.
Data Analysis
Part of my postdoc involved training this amazing and clever team of young scientist on their epidemiological analysis using R/Rstudio and generalized mixed effects models to identify key risk factors for most of these infectious diseases. Most of the training was done remotely (Scotland-Tanzania) due to the COVID-19 pandemic travel restrictions. In summary, the models were developed using the glmmTMB R package as well as other R Epi packages, and ggplot2 for exploratory visualisations. Details on the GLMMs models and risk factors for Leptospira interrogans serovar Hardjo, Coxiella Burnetii, and Brucella abortus can be found in the students published papers.
The other side of my postdoc was analysing this Tanzanian cattle population genotype data to identify genomic regions associated antibody response to these seven pathogens using genome-wide association studies. Different analytical pipelines were used for genotype imputation (e.g., mimimac3, eagle), relatedness, population structure (e.g., Plink), heritability analysis (e.g., ASREML) and GWAS (e.g., GEMMA). This analytical work is in the process of being published (Link to detailed paper will be shared soon) and it was possible with the valuable advice from scientists based at the Roslin Institute - The University of Edinburgh, SRUC, ILRI in Kenya and Tanzania, NM-AIST and more.
Conclusion
This project/role was one of the most amazing experiences of my life as I had the opportunity to visit Africa for the first time, work with amazing scientist and develop my advanced analytical skills to answer epidemiological and genetic scientific questions. The project provided knowledge of the infectious diseases circulating in this important cattle population as well as the key risk factors. It also provides insights into the genetic basis of antibody response to these pathogens.






Servicios
Asesoramiento experto sobre enfermedades infecciosas y transmitidas por vectores, análisis avanzados y visualización de datos para clientes del sector académico, sanitario y de empresas de salud animal.
enriqhernandez18@gmail.com
+44(0) 7452992678
© 2024. All rights reserved.
